Immerse yourself
in origin of human proteome interaction

Human Protein Reference Database
Gene Ontology Consortium
Oncomine Cancer Microarray Database
Interpro
Kinase Pathway Database

Password
forgot password?
About HiMap What's New?
HiMAP is a dynamic browser for the human protein-protein interaction map. HiMAP allows users to begin with a single protein or a set of proteins and explore both known and predicted protein-protein interactions. Literature-confirmed interactions come from the Human Protein Reference Database, yeast-two-hybrid-defined interactions come from two recent publications in Nature and Cell, and predicted interactions were generated by a Bayesian Analysis published in Nature Biotechnology.
HiMAP manuscript published in Nature Biotechnology
HiMap Featured on Nature Signalling Gateway

Science fiction
Discover unknown universes
Biographies
Meet great people
Horror
Enjoy your fears
Romance
Read the best love stories
Once upon a time...
Interaction Map
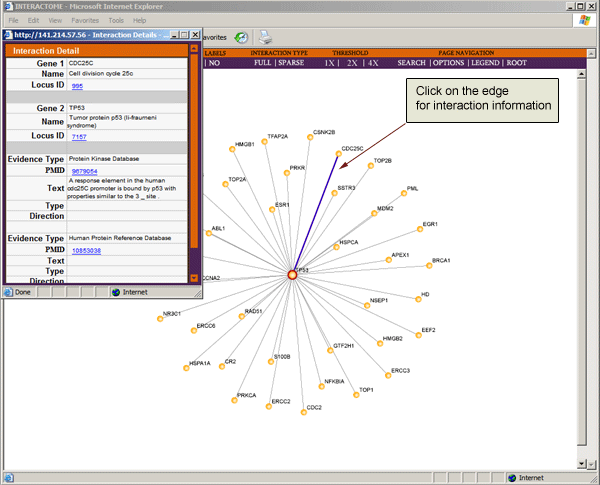
Click on edges to view the literature evidence for a known interaction or the genomic and proteomic evidence for a predicted interaction.
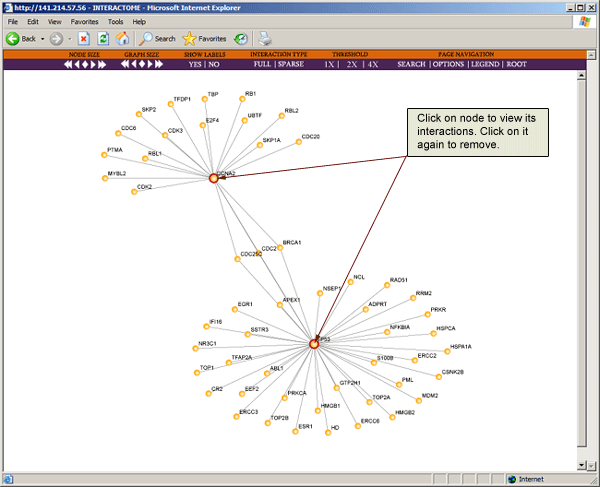
To view all interactions for a particular protein, click on the node to expand the map. You will see that the expanded node will now have a red circle around it. To remove the interactions of the expanded node, just click on it again.
Browser Toolbar
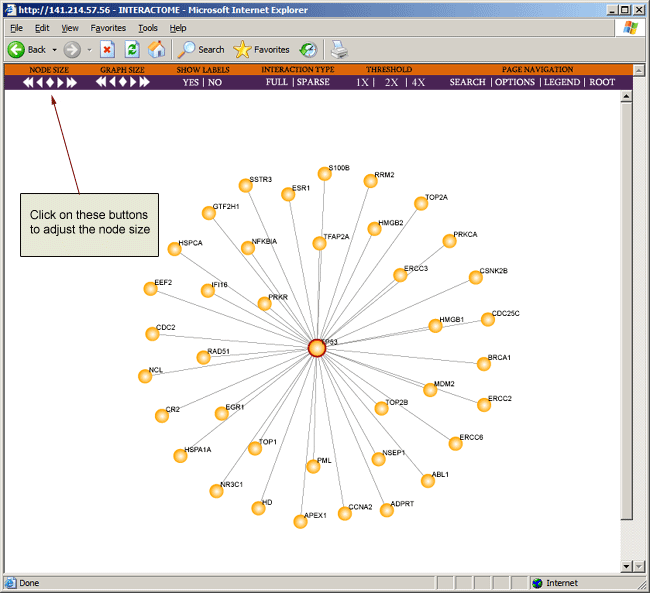
To increase or decrease the size of the node, you can click on the node size buttons on the toolbar located at the top of the browser page.
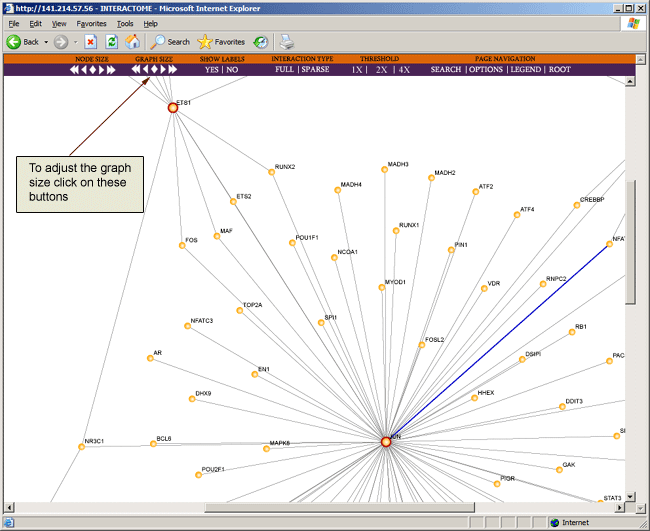
As the interaction map grows larger and more complex, the window may become cluttered and may make veiwing the map difficult. Hence you may wish to adjust the size of the graph by clicking on the graph size buttons on the toolbar located at the top of the browser page.
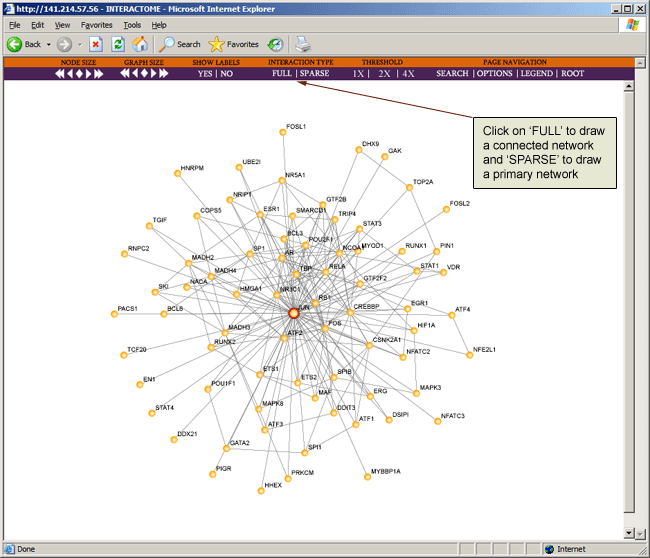
Interaction type "SPARSE" shows primary interactions for only selected nodes (designated with outer red circle). "FULL" shows sparse interactions and all other interactions that connect two nodes in the sparse map.
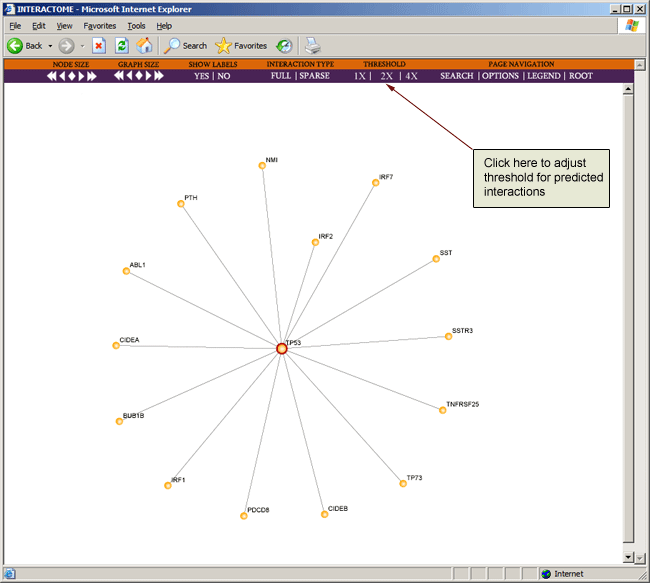
The "Threshold" option corresponds only to predicted interactions and allows users to display interactions at a given confidence level. 1X, 2X, and 4X refer to the posterior odds of the predicted interaction (the default is 2X). Respective likelihood ratios are 381, 762, and 1524
Discover our
limited editions
Explore our top-end Collector's Editions.
Signed and limited tributes to some of the most outstanding writers of all time.






